Andrew C. Calderon
Developing Radioisotope  Responsive Core-Shell Nanoparticle Platforms for Scintillation Proximity Assays (SPA)
Responsive Core-Shell Nanoparticle Platforms for Scintillation Proximity Assays (SPA)
Research:
Physiological analytes in biological systems have traditionally relied upon optical or electrochemical methods for detection. My research however is focused on imaging important signaling molecules such as glucose and inositol phosphates, which are important in cellular signaling pathways and may provide insight into human diseases and disorders such as diabetes. Neither molecule is optically or electrochemically active. Addition of traditional fluorescent or electrochemical labels introduces large moieties compared to these small molecules, possibly affecting their binding properties. As such, we are using 3H as a radiolabel to circumvent this problem as it has minimal effect on the size and binding characteristics of the analyte, having minimal perturbation on the fate of the analyte inside cells.
The goal of my research is to develop nanoparticles that utilize scintillation proximity assay (SPA) to temporally and spatially detect these 3H-labeled analytes in cellular systems. SPA uses a solid-state platform (SPA particle) to absorb energy from radioactive decay events occurring in close proximity to the platform. Energy from radioactive decay events are converted into bursts of photons and detected using photon counters or high sensitivity cameras.1
We are designing nanometer-sized SPA particles (nanoSPA) functionalized with recognition elements (e.g. proteins, aptamers, etc.) for selective detection of radiolabeled analytes. SPA provides selectivity and low background, making the use of nanoSPA advantageous for continuous and separation-free measurements1,2 in cellular systems. Thus far, we have prepared nanoSPA agents for model binding systems (NeutrAvidin-biotin and ds-DNA hybridization) and are extending the technology to other analytes of interest.
Reference
- Handbook of Radioactivity Analysis; L’Annunziata, M.F., Ed.; Academic Press: San Diego, CA, 2003.
- Stockinger, W.; Castoreno, A.B.; Wang, Y.; Pangnon, J.C.; and Nohturfft, A. Real-time analysis of endosomal lipid transport by live cell scintillation proximity assay. J. Lipid Res. 2004, 45, 2151-2158.
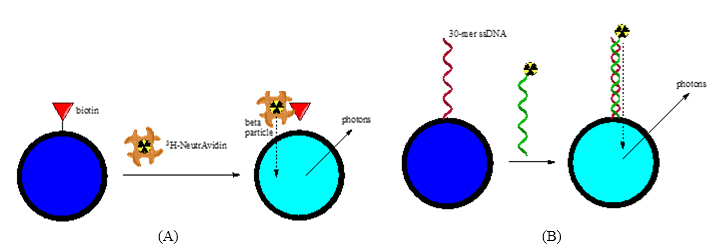
Figure 1. Successful nanoSPA Particle-based Detection of 3H-labeled Analytes through
(A) NeutrAvidin-biotin and (B) DNA Hybridization Binding Models

